Description
Get more out of CRISPR with new insertion protocol

Channabasavaiah Gurumurthy, PhD
CRISPR, the newest gene editing technology, efficiently slices DNA unlike any other known procedure. It’s a scientist’s dream to delete mutated or faulty DNA. But CRISPR solves only half the problem.
While CRISPR can delete DNA, the process for replacing that piece has been unwieldy, imprecise and inefficient.
Easi-CRISPR is a simple protocol that inserts long, single-stranded DNA with remarkably high efficiency rates—up to 100 percent in some genes.
As a research tool, Easi-CRISPR could have a profound impact in developing new genetically engineered model organisms—including animals, cells and plants—and other testing media that could lead to future cures of genetic conditions. The protocol takes approximately two months to generate either knock-in or conditional knock-out mouse models.
To discuss licensing opportunities about this technology, contact Amanda Hawley, PhD, at
ahawley@unmc.edu or 402-310-5602.
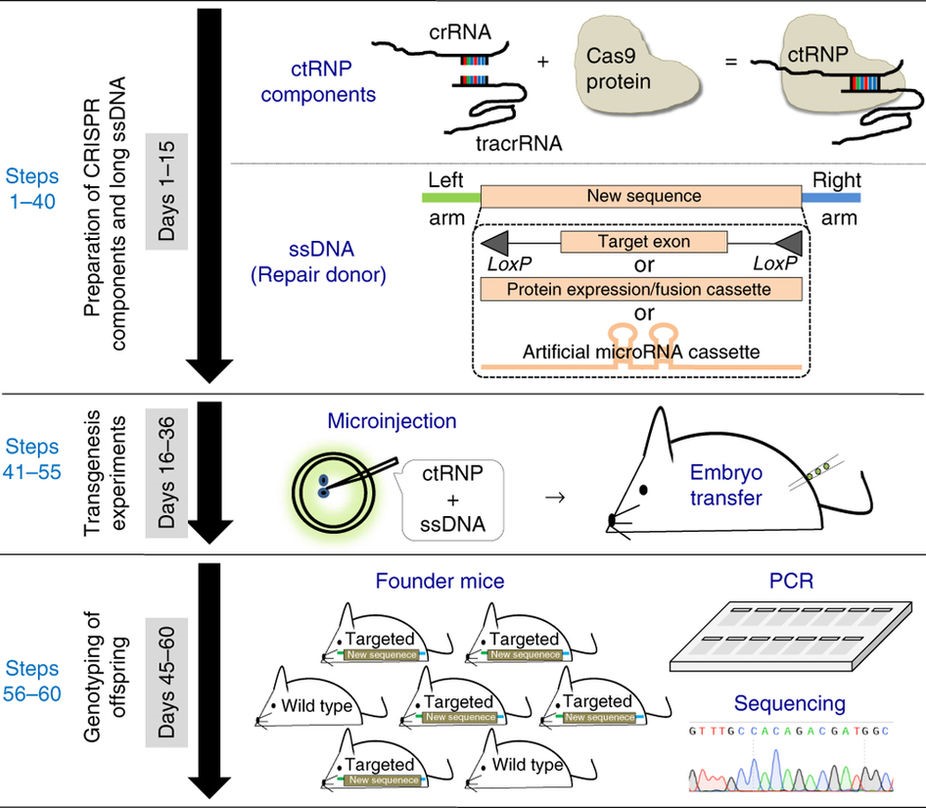
Schematic of Easi-CRISPR
Technical details
CRISPR homologous recombination
Researchers have successfully developed a protocol to simplify the process of generating genetically engineered animal models.
The Easi-CRISPR protocol involves the insertion of long, single-stranded DNA (ssDNA)–up to 1.5 kilobases—which express various types of proteins. Long ssDNA dramatically improves the efficiency of homologous recombination-based targeted insertion. In experiments, the protocol exhibited up to 83 percent insertion efficiency when the ssDNA was ~0.5 kilobases long.
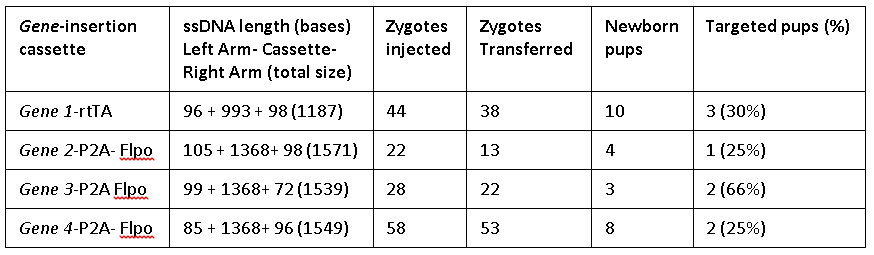
The Easi-CRISPR method inserts ssDNA that ranged from 1.2 to 1.57 kilobases either at the 5’ end or 3’ end, for two distinct genes generating knock-in animal models. The ratios of the exogenous sequence length to homology arm length ranged between 5:1 and 50:1. Efficiency of targeted insertion ranged from 25 percent to 66 percent.
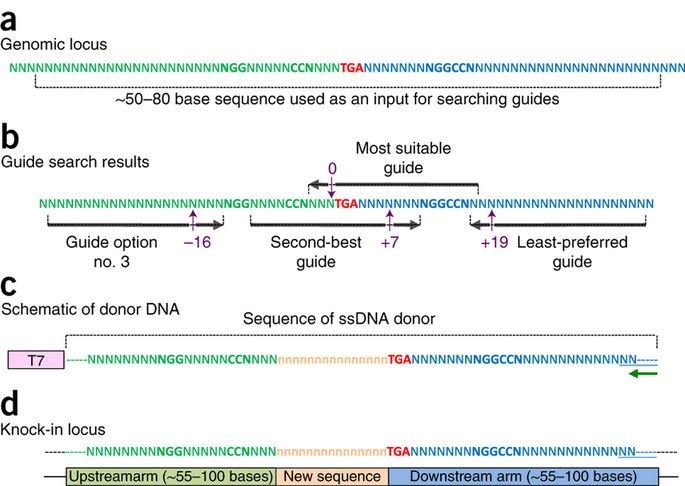
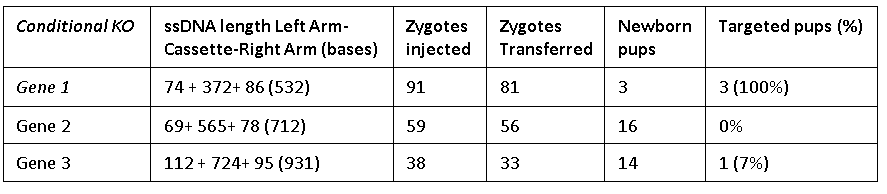
The Easi-CRISPR method inserts a floxed version of ssDNA into mouse zygotes to generate conditional knock-out animal models. The ssDNA cassettes from 532 to 931 bases are shown. Correct insertion of full length cassettes were observed in Gene 1 and Gene 2 (7 percent to 100 percent efficiency). However, some of the Gene 3 samples only contained partial cassette insertion and were scored negative (for full length cassette insertion).
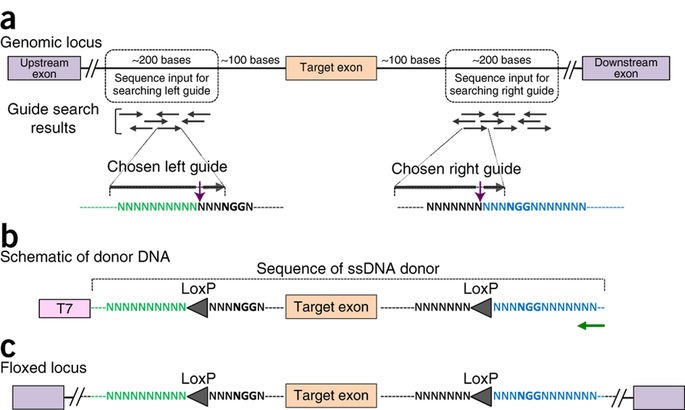
Generating knock-in and conditional knock-out mouse models is a laborious and time-consuming process, sometimes taking a year or more to create fully developed transgenic animals. The Easi-CRISPR approach however, could reduce it to just 2-3 months. The protocol works for any variety of transgenes, requires no special equipment and makes possible genetic editing on an entirely new scale: editing hundreds of base pairs with higher efficiency.
Easi-CRISPR: Microinjection data for floxed allele generation at seven loci
| Gene-insertion cassette | ssDNA length Left Arm-Cassette-Right Arm (bases) [source of ssDNA] | Zygotes injected | Zygotes transferred | Live-born animals (% of transferred zygotes) | Targeted animals (%)a |
| Pitx1-exon 2 floxedb | 93 + 862 + 91
[IvTRT]
| 85 | 76 | 10 (13.2) | 4 (40%)b |
| Ambra1-exon 4 floxed | 96+589+103
[IvTRT]
| 67 | 63 | 8 (12.7) | 6 (75%)c |
| Col12a1-exon 2 floxed | 55 + 527 + 55
[IvTRT]
| 105 | 79 | 3 (3.8) | 3 (100%)d |
| Ubr5-exon 58 floxed | 78+535+86
[IvTRT] | 20 | 16 | 2 (12.1) | 2 (100%)e |
| Syt1-exon 6 floxed | 75 + 635 + 75
[IDT Megamer™] | 51 | 45 | 8 (17.8) | 1 (12.5%)f |
| Syt9-exon 3 floxed | 87 + 893 + 68
[IDT Megamer™] | 43 | 41 | 12 (29.3) | 1 (8.5%)g |
PPP2r2a-exon 3
floxed
| 95 + 619 + 84
[IDT Megamer™] | 34 | 33 | 3 (9.1) | 3 (100%)h |
a. The alleles that did not contain the inserts were not analyzed for the presence of indels because genotyping assays were mainly designed to identify the targeted-insertion alleles. However, noticeable deletions were observed for some samples (e.g., deletions in the non-targeted alleles).
b. Animals #3, #5, #7 and #8 were heterozygous for both 5’ and 3’ LoxP sites. Animal #5 had a floxed allele with 1 nucleotide insertion mutation at the intronic region, which may not affect function. Animals #2, #9 and #10 had only 5’ LoxP site, and animal #4 had only 3’ LoxP site.
c. Animals #1, #2, #3, #5, #7 and #8 were heterozygous for both the 5’ and 3’ LoxP sites. Animal #7 had a floxed allele with 1bp- insertion mutation in the intronic region, which may not affect function.
d. Animals #1 and #2 were heterozygous for both 5’ and 3’ LoxP sites and they carried deletions in their second allele. Animal #3 was biallelic for both LoxP sites.
e. Animals #1 and #2 were heterozygous for both 5’ and 3’ LoxP sites.
f. Animals #4 and #7 had only 5’ LoxP insertion and the animal #6 had correctly targeted LoxP sites.
g. Animal #12 had correctly targeted LoxP sites and all others were wild type.
h. Animals # 1 and # 2 are heterozygous for both loxPs with deletions in the second allele and the pup #3 is biallelic
Easi-CRISPR: Microinjection data for knock-in allele generation at six loci
| Gene-insertion cassette | ssDNA length Left Arm-Cassette-Right Arm (bases) [Source of ssDNA] | Zygotes injected | Zygotes transferred | Live-born Animals (% of transferred zygotes) | Targeted animals (%)a |
| Fgf8-P2A-FlpO | 105 + 1368 + 98
[IDT Megamer™] | 22 | 13 | 4 (30.8) | 1 (25%)b |
| Slc26a5-P2A-FlpO | 99 + 1368 + 72
[IDT Megamer™]
| 28 | 22 | 3 (13.6) | 1 (33%)c |
| Mafb-P2A-FlpO | 85 + 1368 + 96
[IDT Megamer™]
| 58 | 53 | 8 (15.1) | 2 (25%) |
| Otoa-rtTA | 96 + 1220 + 98
[IDT Megamer™]
| 19 | 18 | 2 (11.1) | 1 (50%) |
| Mmp9-T2A-mCitrine | 60 + 782 + 60
[IvTRT}
| 52 | 50 | 12 (24) | 8 (67%)d |
| Mmp13-T2A-mCherry | 60 + 779 + 60
[IvTRT]
| 55 | 52 | 10 (19.2) | 4 (40%) |
a. The alleles that did not contain the inserts were not analyzed for the presence of indels because genotyping assays were mainly designed to identify the targeted-insertion alleles. However, noticeable sequence additions or deletions were observed for some samples in these assays
b. Animal #4 had bi-allelic insertions of the knock-in cassette.
c. Animal #1 had additional sequences at the 3’ junction (sequence not fully characterized and Pup #3 had a precise insertion at both junctions.
d. Animal #4 appeared to be mosaic containing both a correctly targeted allele and a deletion in the 3’ junction (sequence not fully characterized)